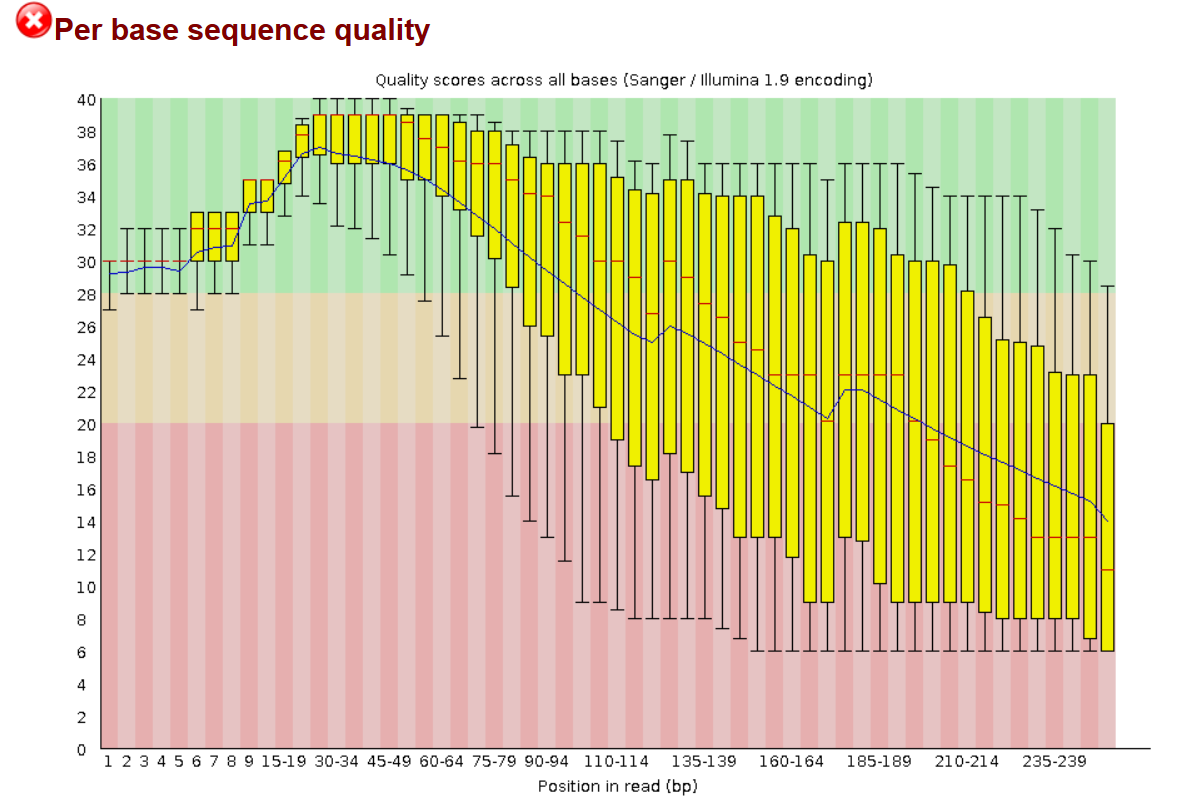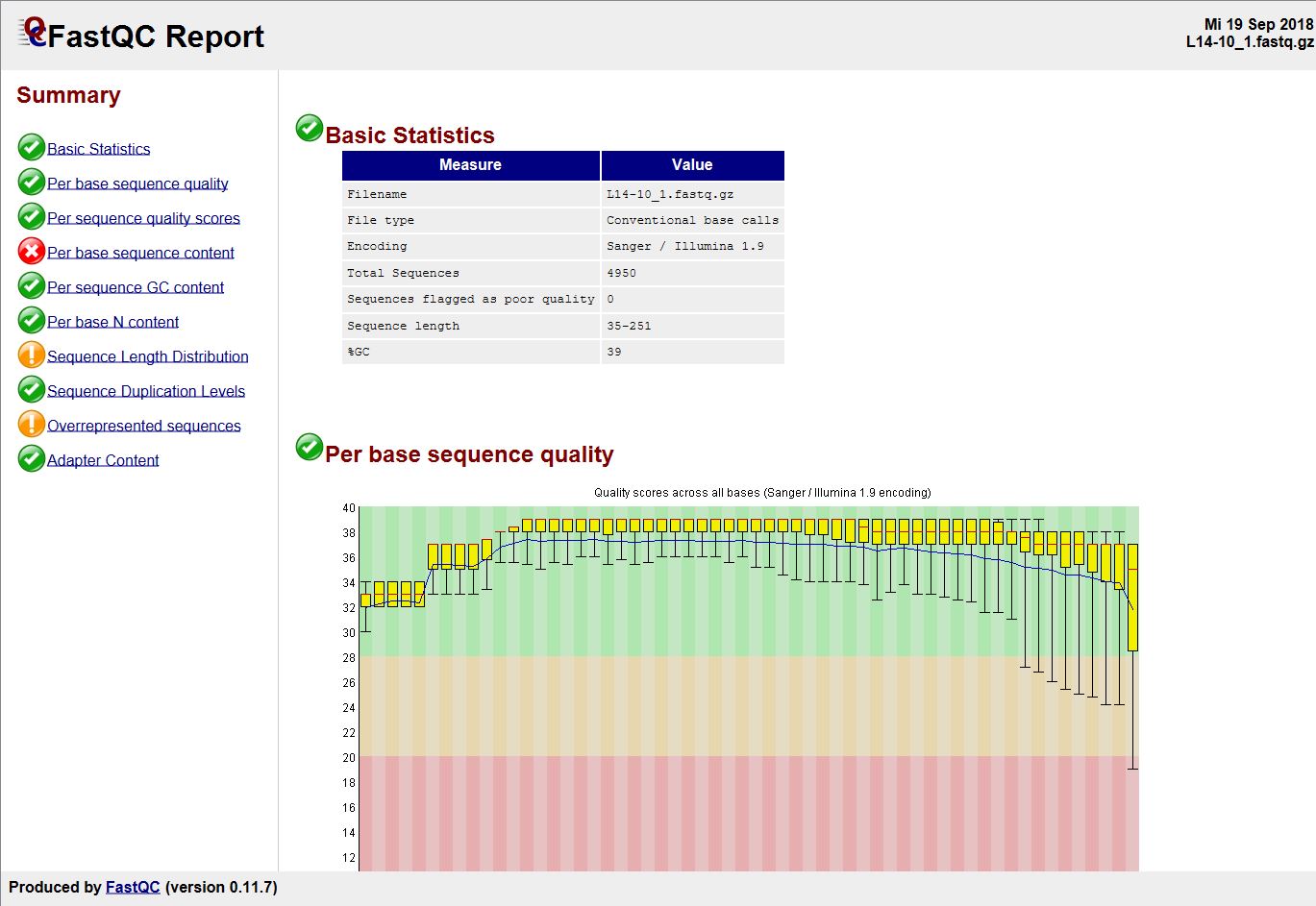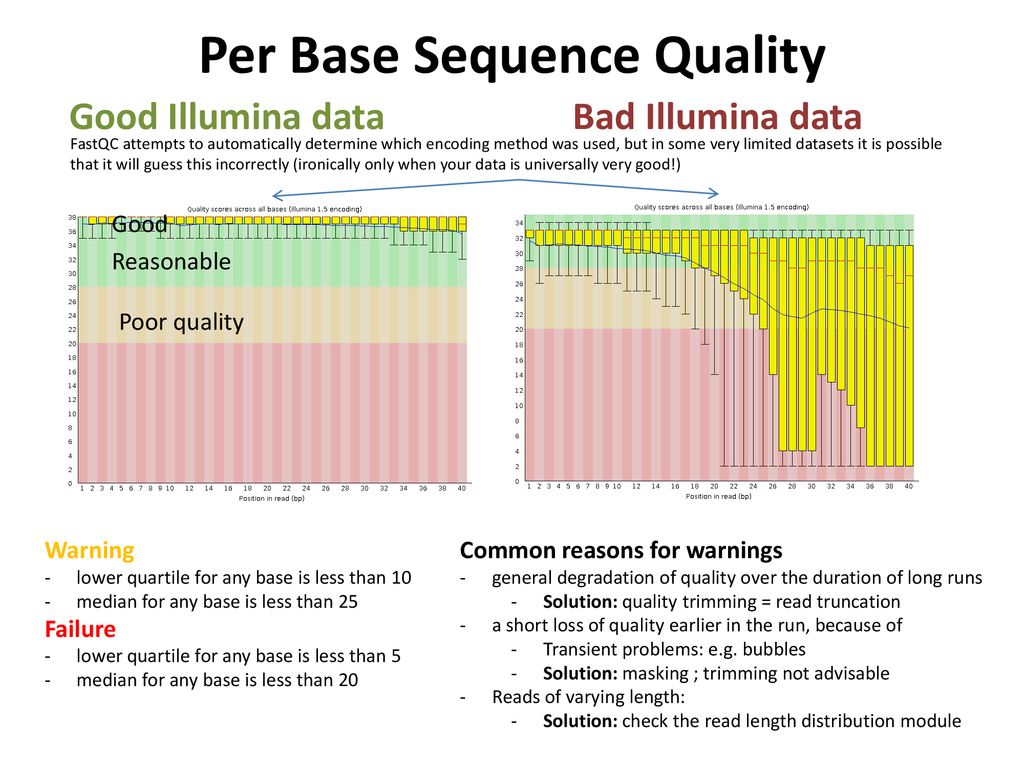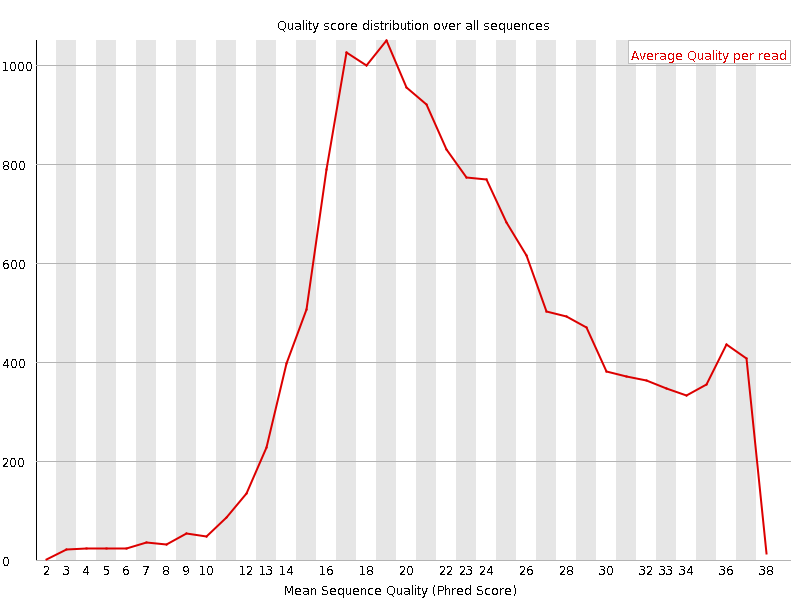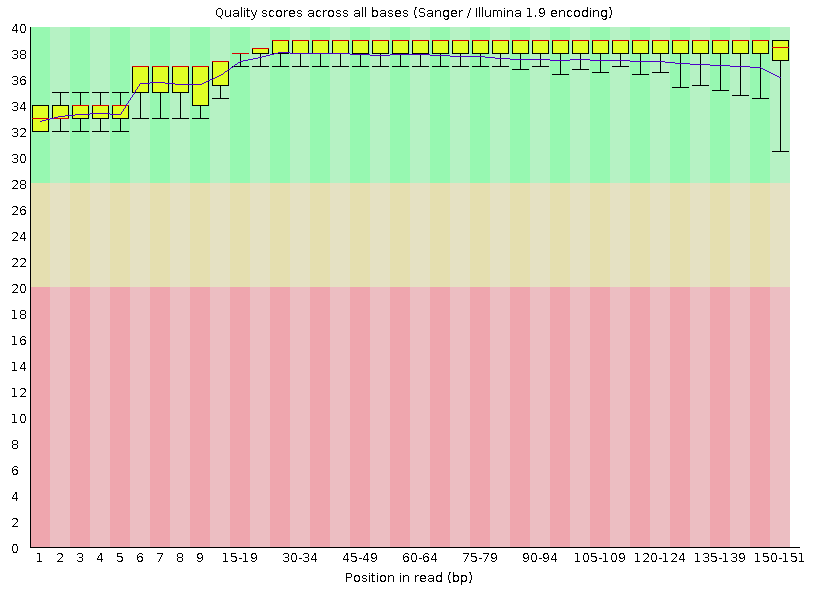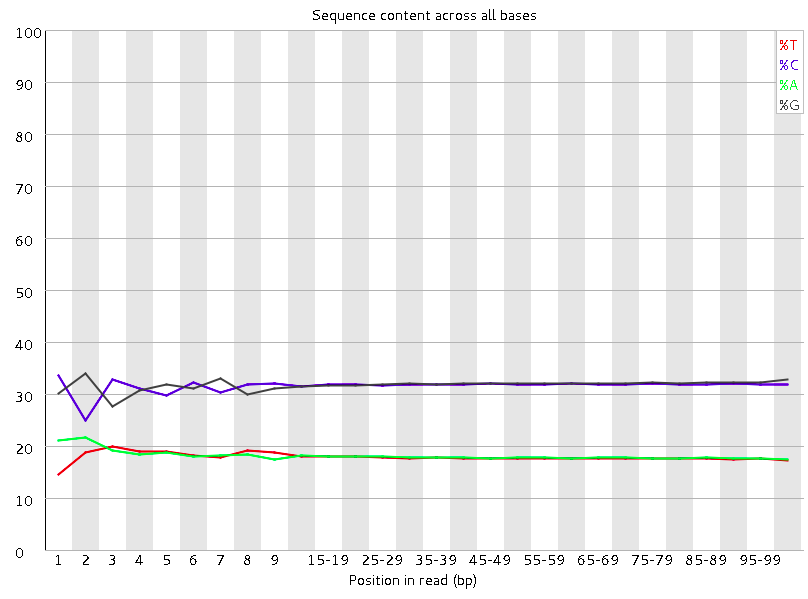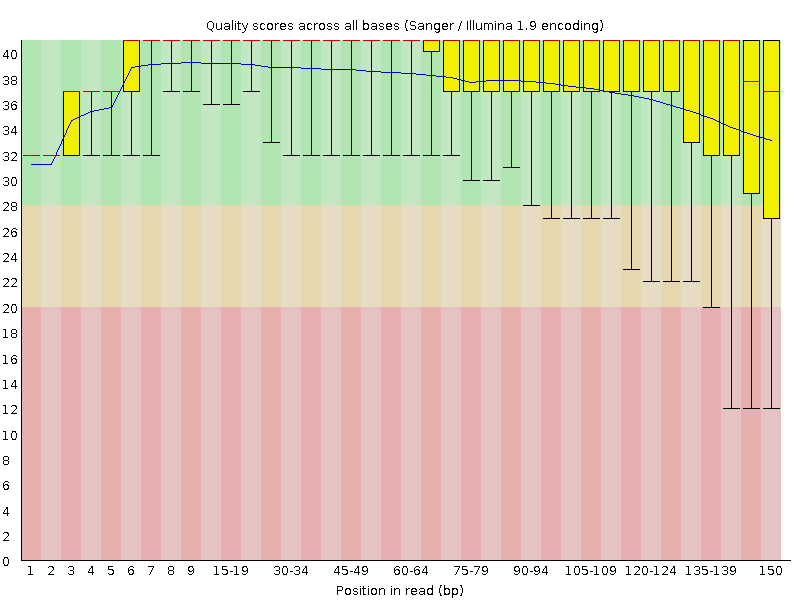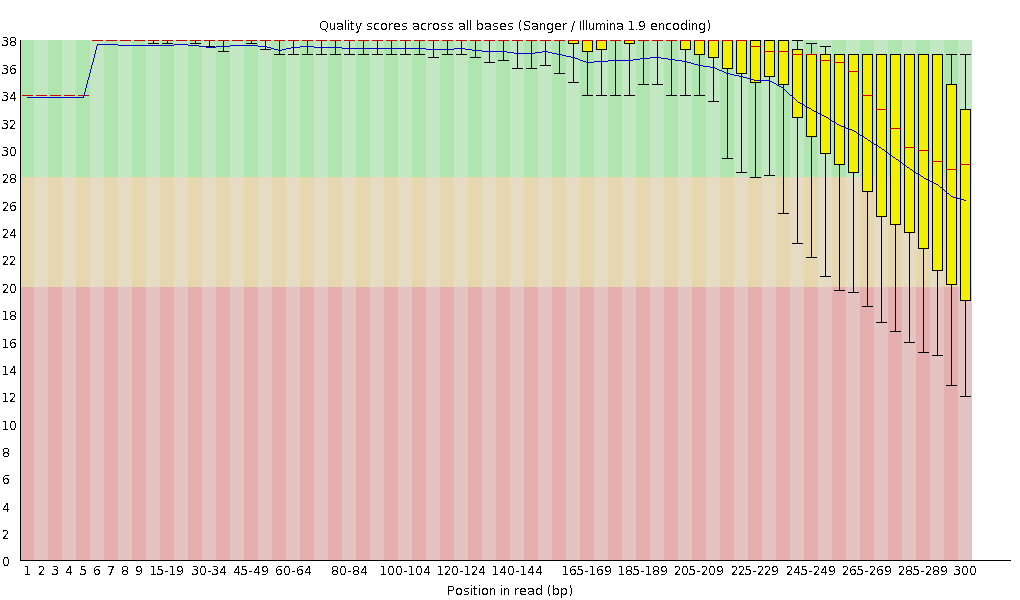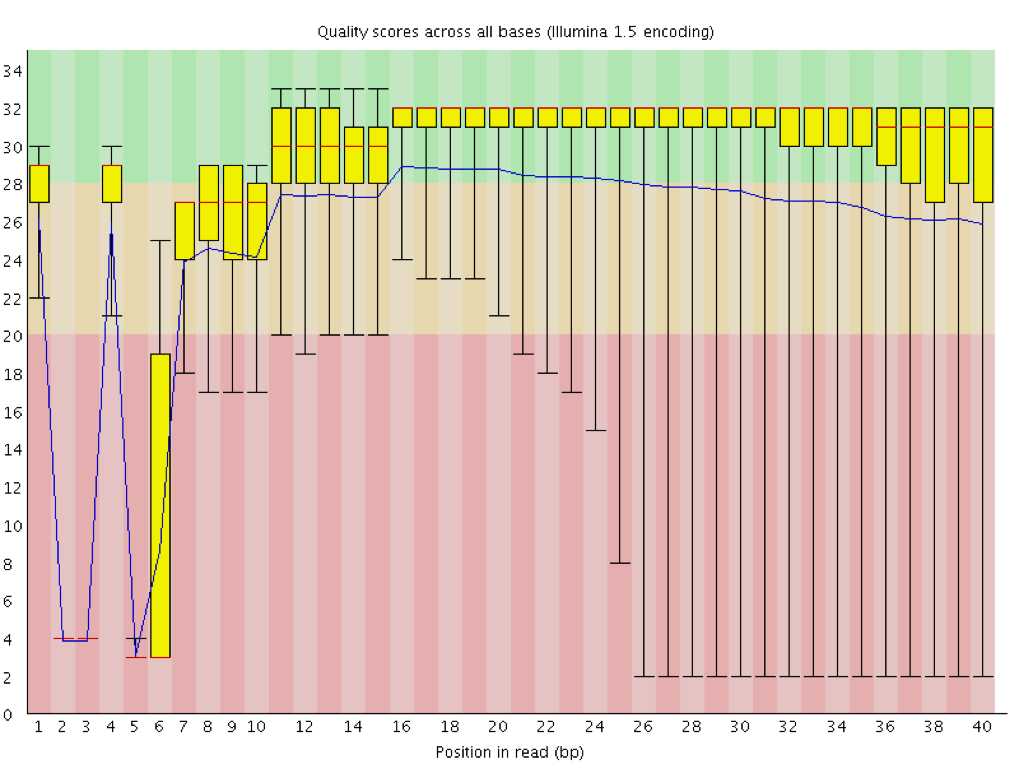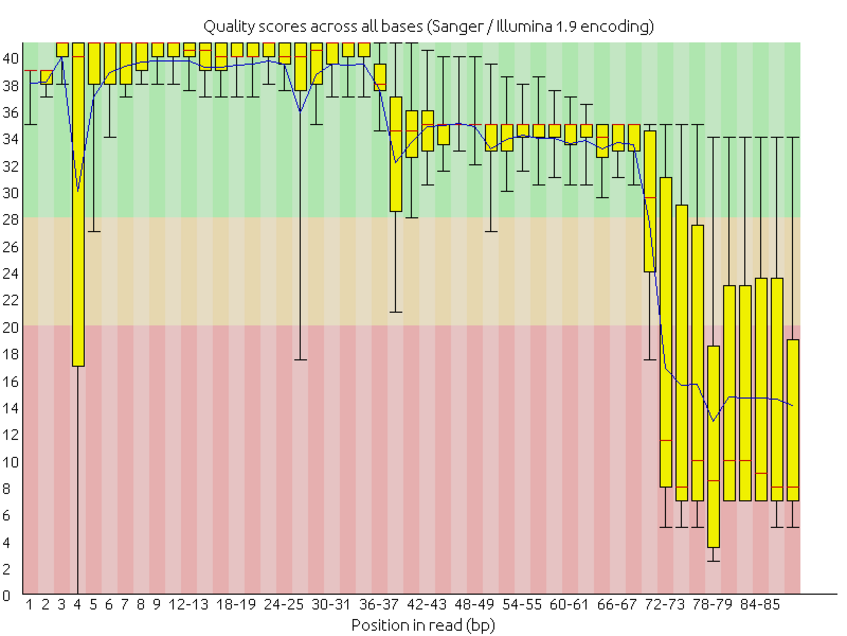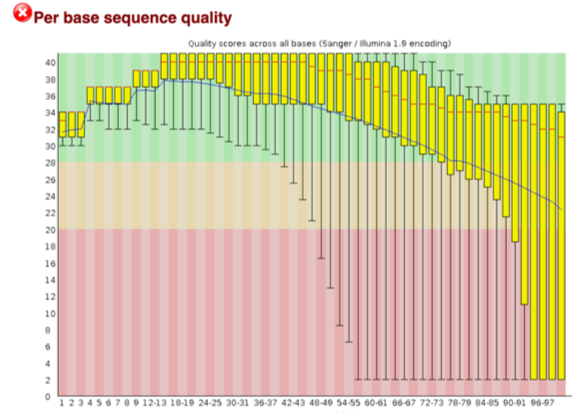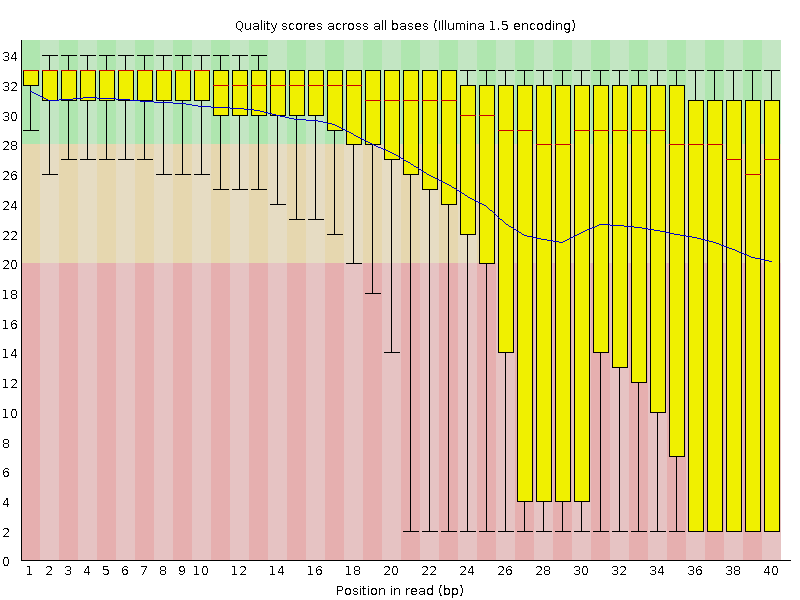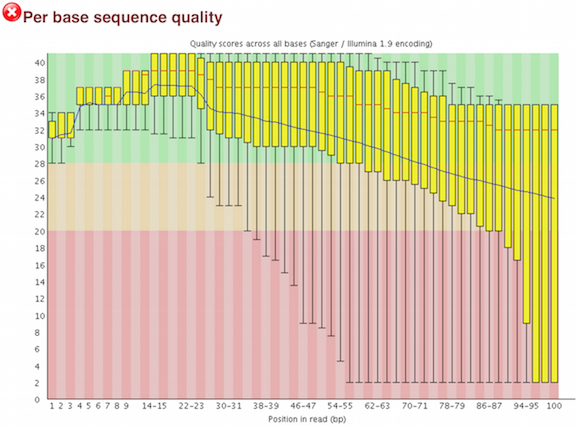
Quality control: Assessing FASTQC results | Introduction to RNA-Seq using high-performance computing - ARCHIVED

SciBear🇺🇦 on Twitter: "(1/7) A quality control tool for raw #sequence data. Using #FastQC you may check: 🚀 Per base sequence #quality (do you see a drop in sequencing quality near the
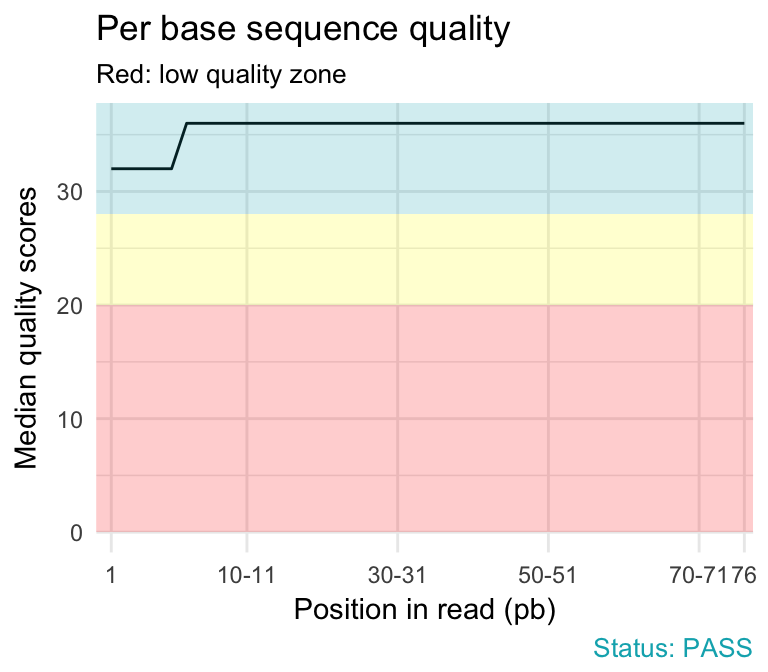
fastqcr: An R Package Facilitating Quality Controls of Sequencing Data for Large Numbers of Samples - Easy Guides - Wiki - STHDA